Research
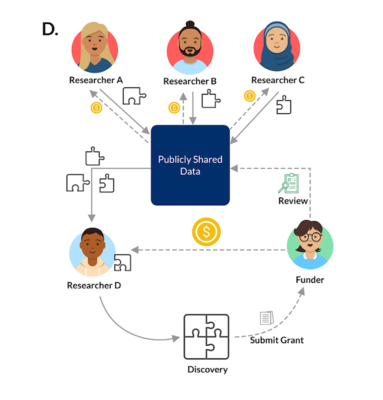
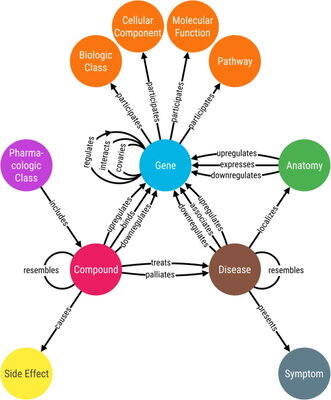
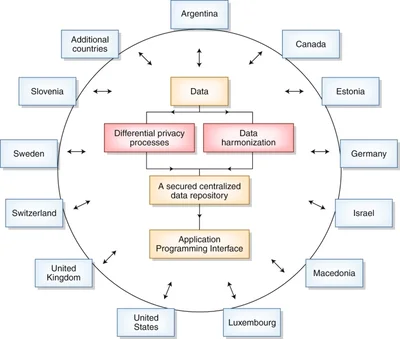
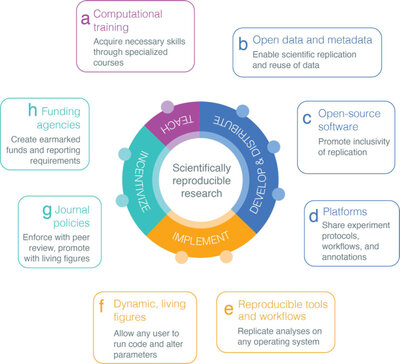
There is an abundance of publicly available data about various biological systems, but it can be difficult to draw insight from individual datasets. Our lab develops algorithms that integrate these data to help model and understand complex biological systems. Doing this allows us to investigate many different biological conditions, including those with limited data, such as rare diseases. We recognize that our lab won’t have all the answers, or even all of the questions, so we aim to develop tools and processes that any biologist can reuse. Our approach to research prioritizes transparency, rigor, and reproducibility.
The citations on this page were generated automatically from just identifiers using the Manubot cite utility developed right here in the Greene Lab!
2026
2025
2024
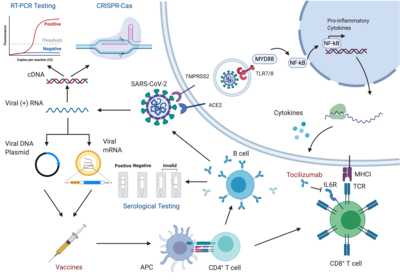
2023
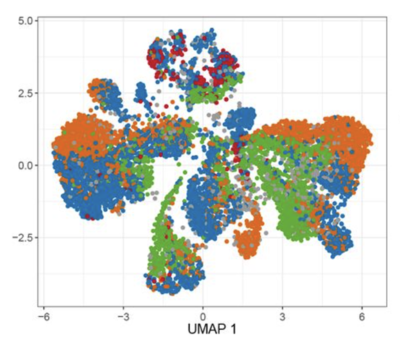
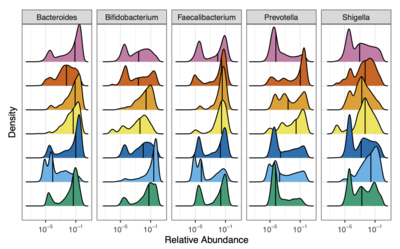

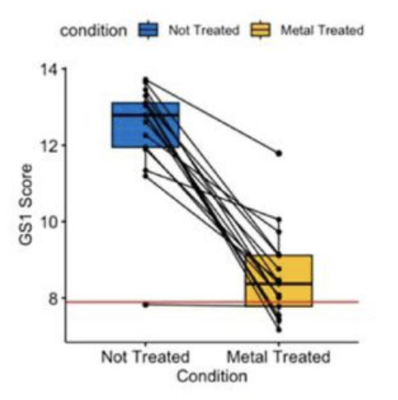

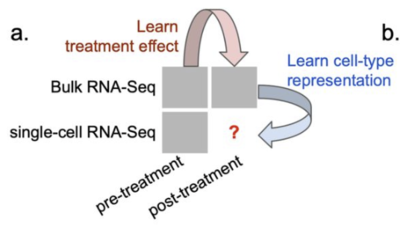

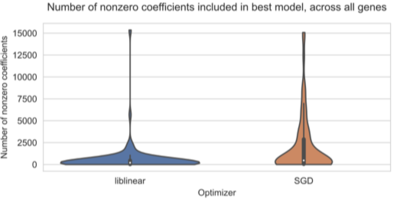
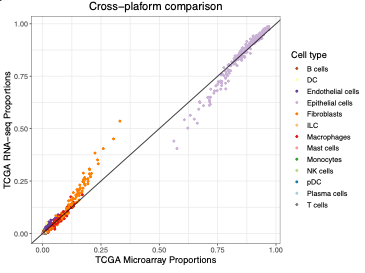
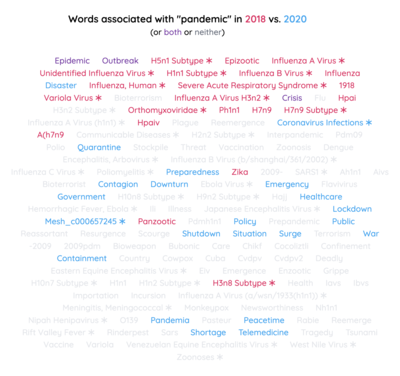


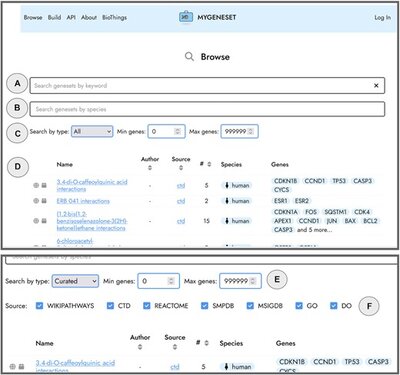
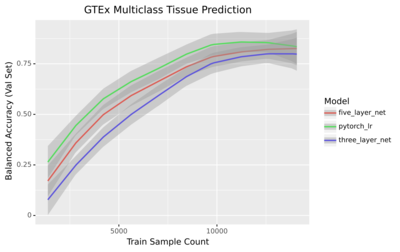
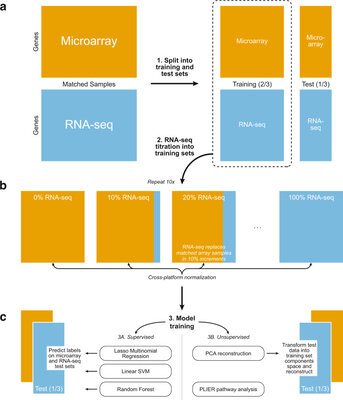
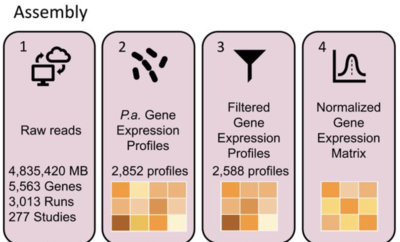

2022
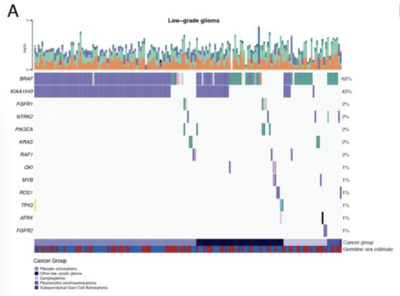
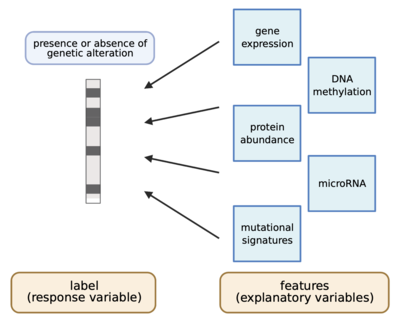
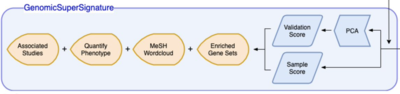






2021





2020

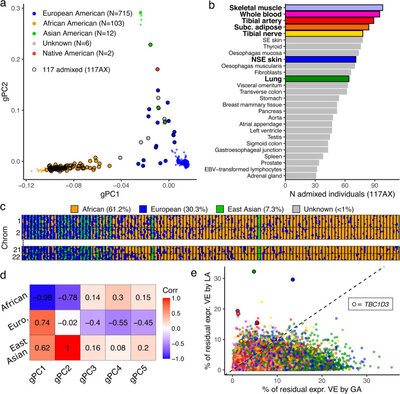
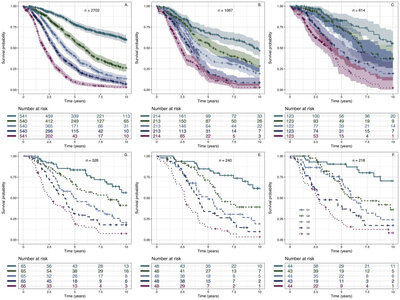
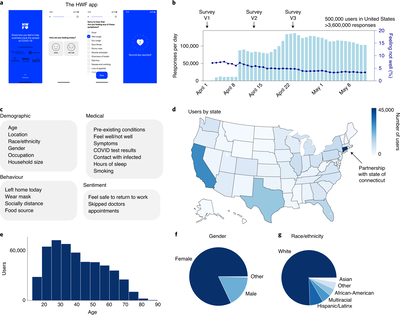
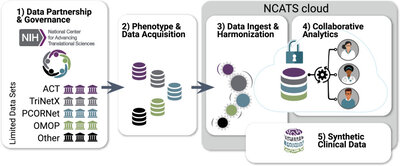
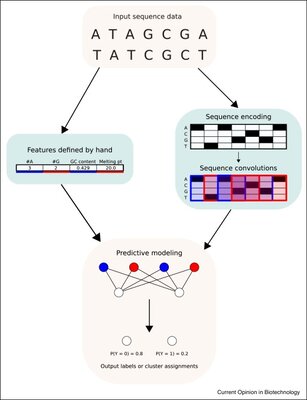
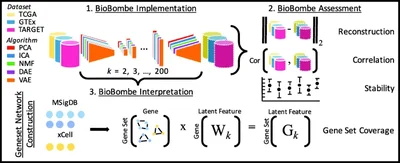
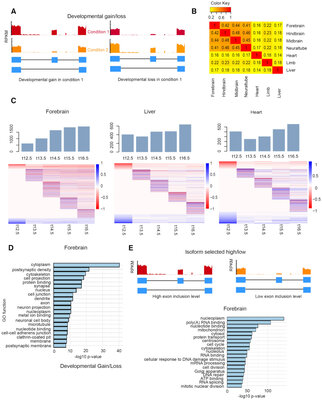
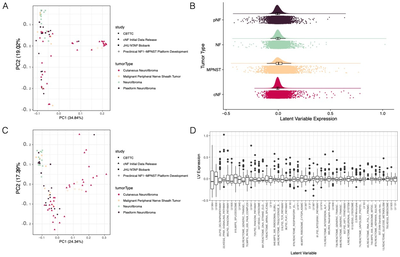
2019





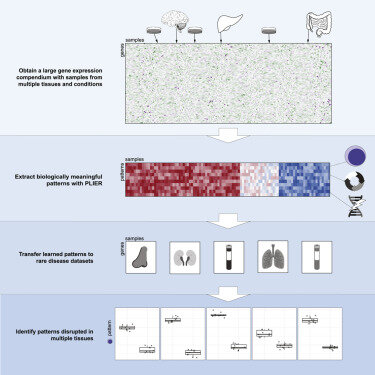



2018
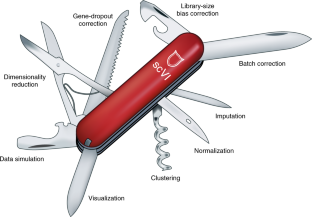

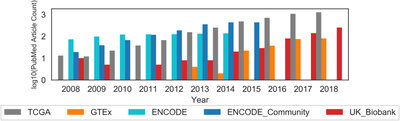



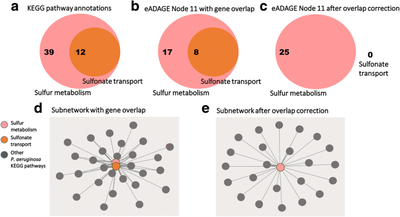
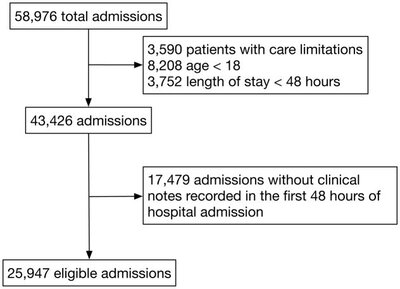




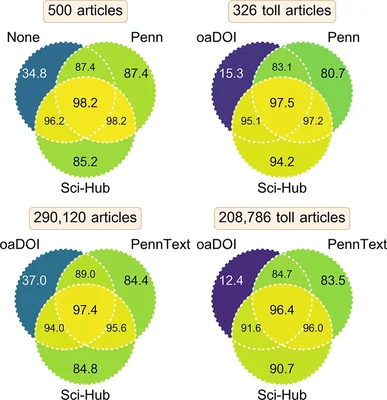


2017
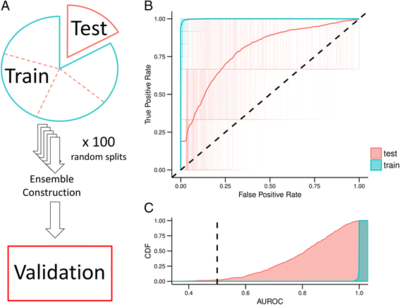




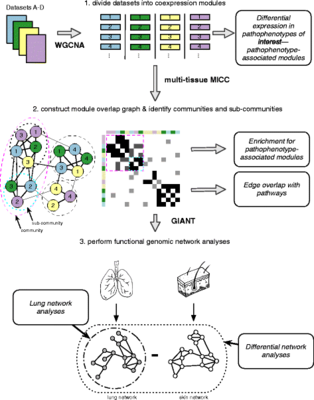

2016
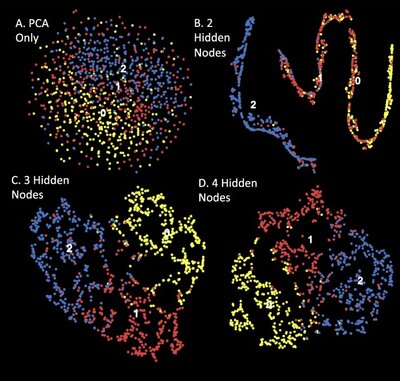


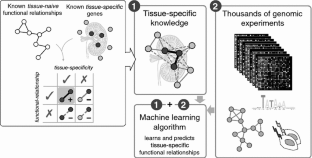
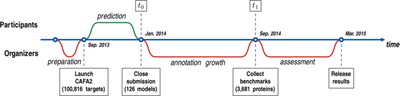
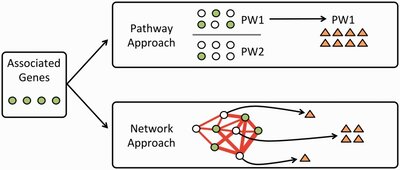
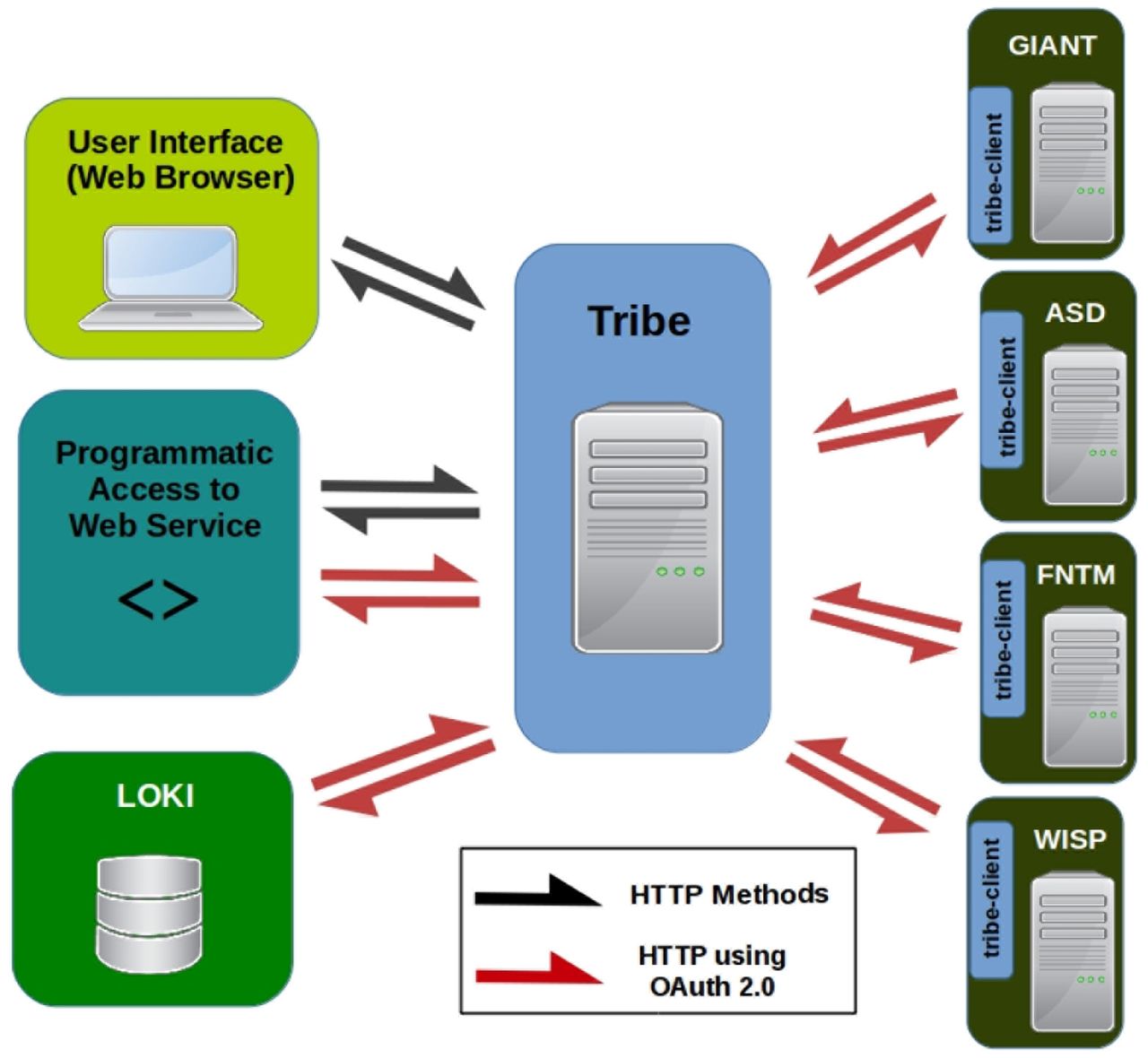

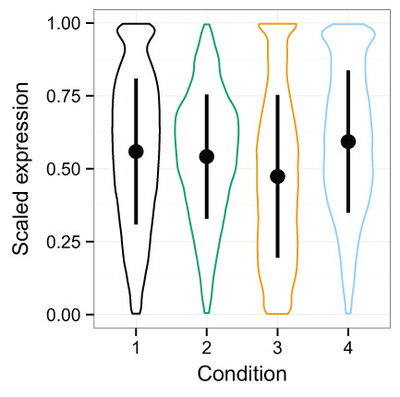

2015


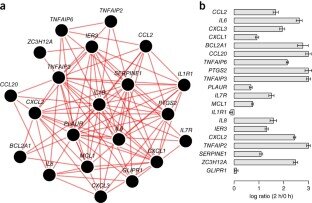
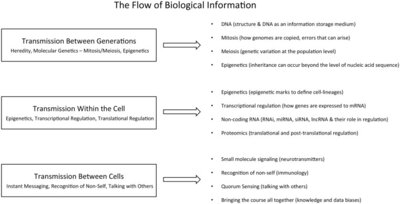
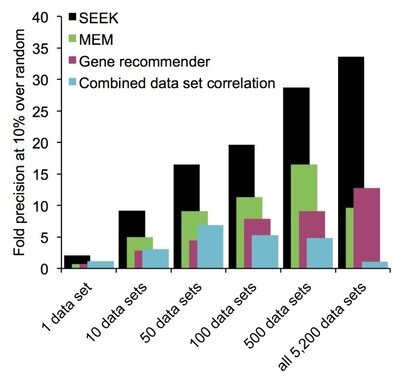
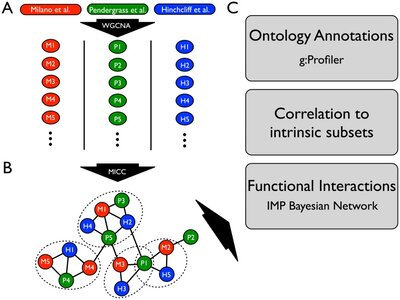

2014
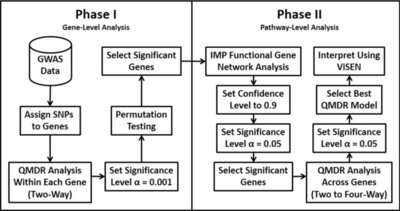
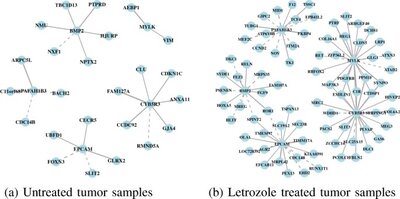
2013
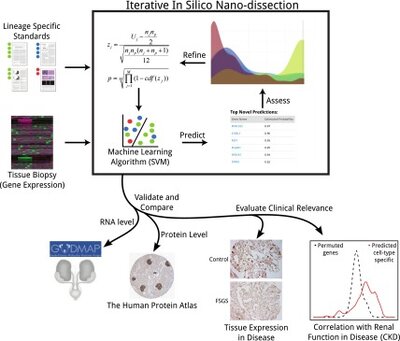
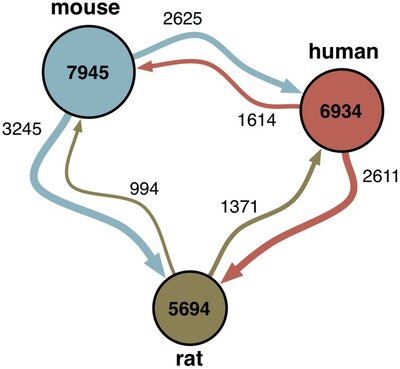

2012